Quantitative Biomarkers and Artificial Intelligence in Musculoskeletal Imaging
Our lab focuses on quantitative imaging of the musculoskeletal systems to identify biomarkers and assess the progression and severity of diseases and pain. Our primary objective is informing clinicians to improve treatment strategies and ultimately enhance the quality of life for patients affected by musculoskeletal conditions. The conditions we focus on are hip and knee joint osteoarthritis and degeneration as well as back pain and injury. To achieve this, we leverage artificial intelligence and develop/employ a range of techniques including MRI sequence development and end-to-end processing of imaging data.
Our lab is led by Professor Sharmila Majumdar and is a part of the Musculoskeletal and Quantitative Imaging Research Group (MQIR) group in the Department of Radiology and Biomedical Imaging at UCSF.
Ongoing Research Projects
KICK: Hip-Knee joint bone and cartilage interaction
Analyzing the complex pathophysiology of knee and hip joint degeneration and impact of gait biomechanics in hip OA patients (KICK-Study)
In this study, we are focusing on the hip-knee joint bone and cartilage interaction and the association of bone shape with incident and severe OA condition and investigate the difference in loading pattern using in-vivo motion analysis tool. Our goal is to evaluate the changes in the bilateral bone shape asymmetry of hip and knee joint and understand the mediation effects of gait biomechanics. This is a longitudinal study investigating 100 people with unilateral hip OA condition.
Related publications
- Thahakoya R et al. Evaluating the relationship of proximal bone shape asymmetry with cartilage health and biomechanics in patients with hip OA. In Proceedings of the 31st Annual Meeting of ISMRM, Toronto, Ontario, Canada, 2023. 4373
- Bhattacharjee R, Luitjens J, Han M, Thahakoya R, Roach K, Souza R, Pedoia V, Majumdar S. Preserved patellar cartilage at the expense of anterior femoral cartilage damage in Hip Osteoarthritis: An Inter-Limb Inter-Joint qMRI analysis. In Proceedings of the 31st Annual Meeting of ISMRM, Toronto, Ontario, Canada, 2023. 2454.
Project members at UCSF
Rafeek Thahakoya, Rupsa Bhattacharjee, Eric Hammond, Misung Han, Emma Bahroos, Valentina Pedoia, Richard Souza, Sharmila Majumdar
Funding number
NIHR01AR069006
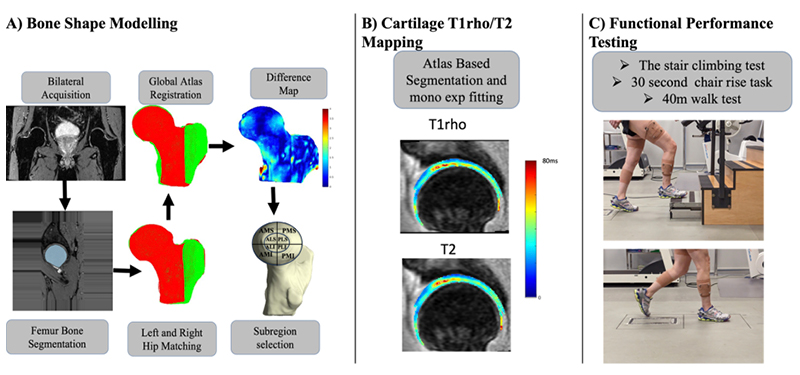
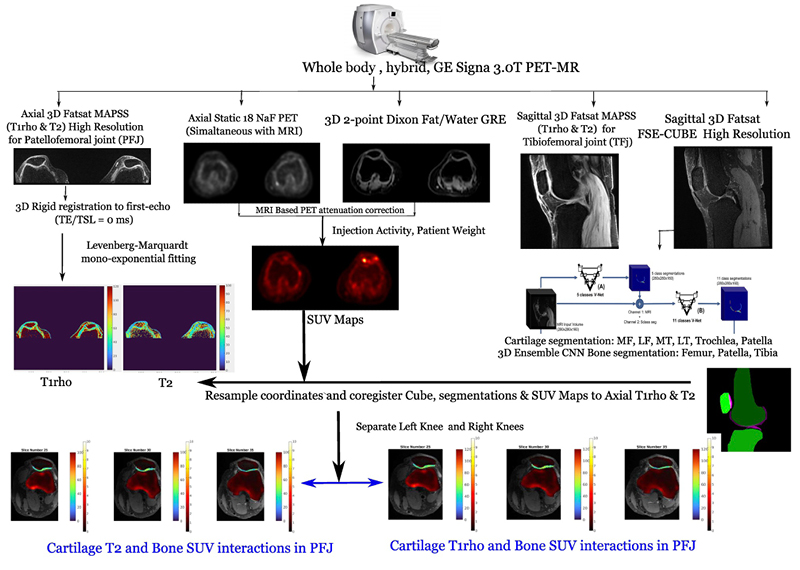
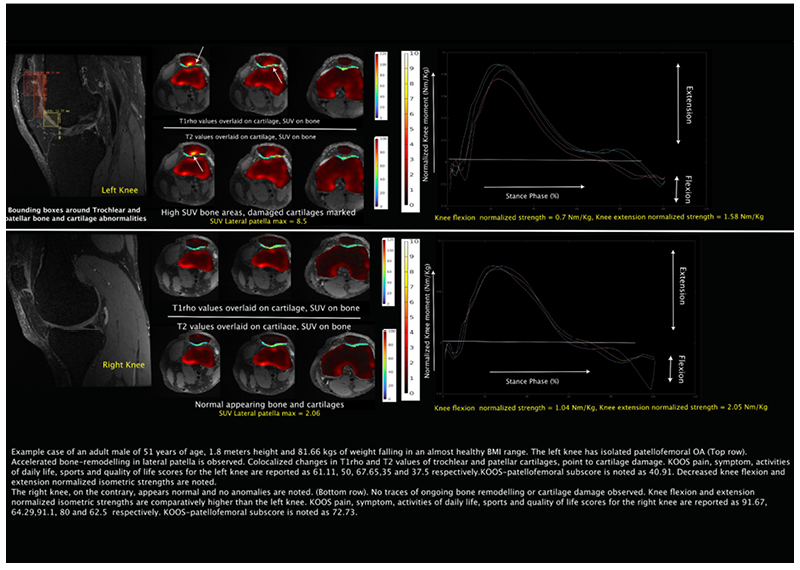
PFJOA: Bone and cartilage interaction of patellofemoral joint
Simultaneous Imaging of Tissue Biochemistry and Metabolism Associated with Biomechanics in Patellofemoral Joint Osteoarthritis (PFJOA)
In this prospective study, we are aiming for a deeper understanding of the mechanics underlying bone and cartilage interactions (via simultaneous bilateral PET-MRI) and the mediation role of gait biomechanics and bone morphology for human subjects with isolated patellofemoral joint osteoarthritis (PFJ-OA). Our overall goal is to: (i) identify cross-sectional and longitudinal local patterns of cartilage and bone interactions unique to PFJOA, and (ii) determine the mediation effects of gait biomechanics and bone morphology on PFJOA progression. We are conducting a longitudinal cohort study investigating 100 people with isolated PFJOA. Simultaneous PET-MRI and gait biomechanics are being collected for all subjects at baseline and 2-year follow-up.
Related publications
- Tibrewala R, Pedoia V, Bucknor M, Majumdar S. Principal Component Analysis of Simultaneous PET-MRI Reveals Patterns of Bone-Cartilage Interactions in Osteoarthritis. Journal of magnetic resonance imaging: JMRI. 2020. Epub 2020/03/25. doi: 10.1002/jmri.27146. PubMed PMID: 32207870.
- Bhattacharjee R, Hammond C, Roach K, Bahroos E, Souza R, Majumdar S, Pedoia V, Towards the understanding of the role of Functional strength markers in Cartilage-Bone Cross Talk: a PET/MRI study in isolated PFJOA patients, ISMRM 2023, Toronto, Canada, (Proc. Intl. Soc. Mag. Reson. Med. 31(2023), Page Nu-0101)
Project members at UCSF
Rupsa Bhattacharjee, Valentina Pedoia, Richard Souza, Sharmila Majumdar, Eric Hammond, Laura Chen, Joshua Johnson, Misung Han, Emma Bahroos
Funding number
NIHR56AR079647
BACPAC: Preventing and Treating Low Back Pain and Disability
BACPAC: Preventing and Treating Low Back Pain and Disability
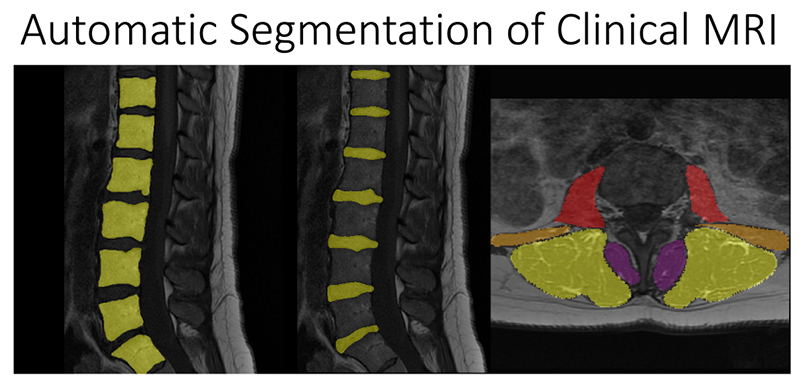
Chronic low back pain is a leading cause of disability in the United States, affecting 1 in 5 adults. Pain sources are nonspecific in almost two thirds of cases, making it difficult for doctors and patients to come up with effective treatment strategies. This lack of effective treatments strategies has led increasing treatment costs to patients and use of opioids to manage pain. Magnetic resonance imaging (MRI) is an effective technique for three-dimensionally imaging the lumbar spine, but imaging findings are notoriously ambiguous indicators due to their lack of consistent associations with clinical symptoms. National Institute of Health’s Back Pain Consortium Research Program (BACPAC) aims to develop our scientific understanding of the driving causes of chronic low back pain, so that we can create better, curative, individualized treatment strategies for patients and reduce opioid use.
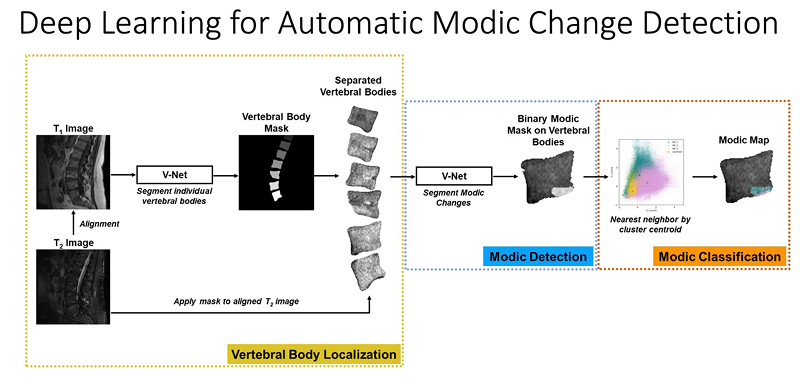
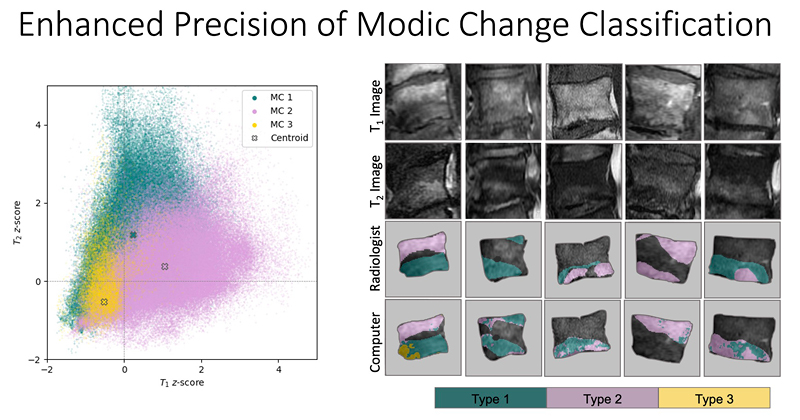
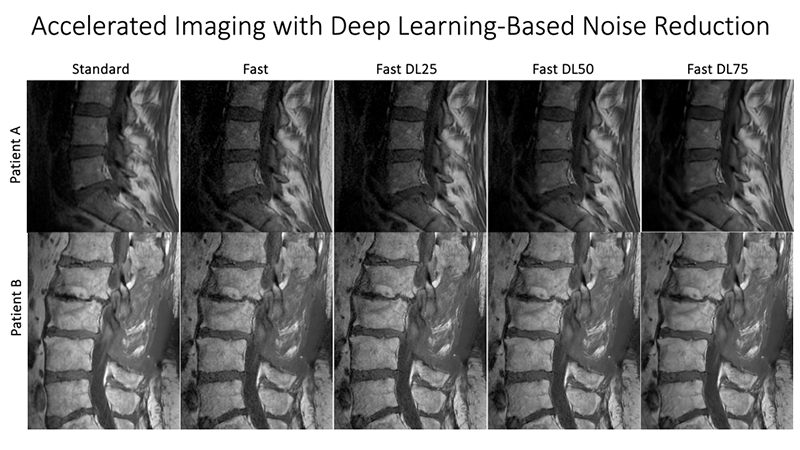
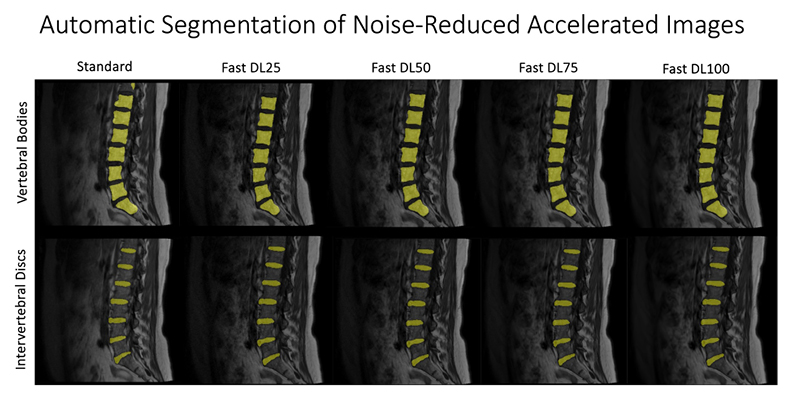
At UCSF ci2, we are contributing to the consortium by collecting and analyzing MR images of people with chronic low back pain to learn how we can use quantitative image-based findings to learn about and treat clinical symptoms. We also build tools to automate quantitative image analysis for clinical use to improve routine patient care and outcomes, including automatic bone, disc and muscle segmentation and characterization. We have trained deep learning-based models to detect anomalies like modic changes, stenotic narrowing in the central spinal canal, vertebral body fractures, and intervertebral disc compressions from MRI. These models have been demonstrated as useful assistants in educating and improving performance in trainees. Scaling these models has helped both our researchers and our collaborators identify new findings and associations between quantitative image-based metrics and chronic low back pain by enhancing dataset size. To put these findings into practice, we deployed those models inline with image acquisition for testing in real time. We hope to translate this work to the clinic and someday empower treating physicians can with AI-assisted data enrichment to develop more effective, individualized interventions to help patients stay happier and healthier.
Related Publications
- Bharadwaj, Upasana Upadhyay, Miranda Christine, Steven Li, Dean Chou, Valentina Pedoia, Thomas M. Link, Cynthia T. Chin, and Sharmila Majumdar. “Deep Learning for Automated, Interpretable Classification of Lumbar Spinal Stenosis and Facet Arthropathy from Axial MRI.” European Radiology 33, no. 5 (May 1, 2023): 3435–43. https://doi.org/10.1007/s00330-023-09483-6.
- Gao, Kenneth T., Radhika Tibrewala, Madeline Hess, Upasana U. Bharadwaj, Gaurav Inamdar, Thomas M. Link, Cynthia T. Chin, Valentina Pedoia, and Sharmila Majumdar. “Automatic Detection and Voxel-Wise Mapping of Lumbar Spine Modic Changes with Deep Learning.” JOR SPINE 5, no. 2 (2022): e1204. https://doi.org/10.1002/jsp2.1204.
- Han, Misung, Emma Bahroos, Madeline E Hess, Cynthia T Chin, Kenneth T Gao, David D Shin, Javier E Villanueva-Meyer, Thomas M Link, Valentina Pedoia, and Sharmila Majumdar. “Technology and Tool Development for BACPAC: Qualitative and Quantitative Analysis of Accelerated Lumbar Spine MRI with Deep-Learning Based Image Reconstruction at 3T.” Pain Medicine 24, no. Supplement_1 (August 1, 2023): S149–59. https://doi.org/10.1093/pm/pnad035.
- Hess, Madeline, Brett Allaire, Kenneth T Gao, Radhika Tibrewala, Gaurav Inamdar, Upasana Bharadwaj, Cynthia Chin, et al. “Deep Learning for Multi-Tissue Segmentation and Fully Automatic Personalized Biomechanical Models from BACPAC Clinical Lumbar Spine MRI.” Pain Medicine 24, no. Supplement_1 (August 1, 2023): S139–48. https://doi.org/10.1093/pm/pnac142.
Project members at UCSF
Sharmila Majumdar, Misung Han, Felix Liu, Christine Park, Eugene Ozhinsky, Michelle Tong, Jennifer Cummings, Isabelle Remick, Emma Bahroos, Madeline Hess, Cynthia Chin, Mark Choe, Upasana Bharadwaj, Valentina Pedoia, Kenneth Gao
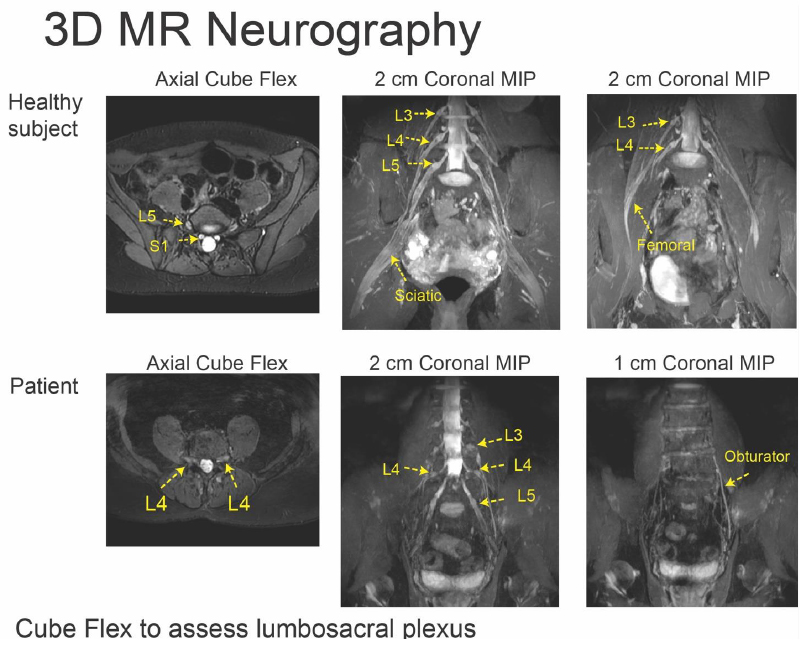
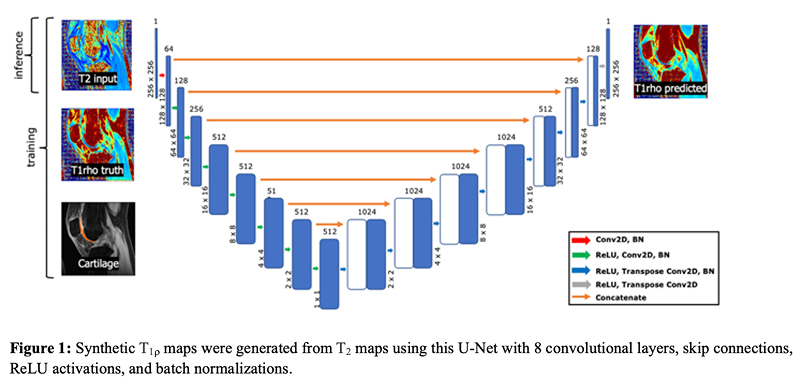
Generating Synthetic T1ρ Maps from T2 maps for Knee MRI with Deep Learning
Generating Synthetic T1ρ Maps from T2 Maps for Knee MRI with Deep Learning
A 2D U-Net was trained to generate synthetic T1rho maps from T2 maps for knee MRI to explore the feasibility of domain adaptation to enrich existing datasets or for faster image reconstruction. The network was developed using images from two prior research studies across three institutions. Network generalizability was evaluated on two new datasets acquired as part of the standard-of-care acquired in a clinical setting and from simultaneous bilateral acquisition in a research setting. This study found the network performed excellent reconstruction of T1rho maps preserving textures and local T1rho elevation patterns in cartilage with NMSE of 2.4% and Pearson’s correlation coefficient of 0.93. Decreased performance for external datasets may be attributed to slight variation in acquisition from different MR scanners and knee coils, suggesting deep learning networks would benefit from volume-wise consideration of scanner properties for performance agnostic to MR scanner equipment.
Project members at UCSF
Michelle Tong, Aniket Tolpadi, Rupsa Bhattacharjee, Misung Han, Sharmila Majumdar, Valentina Pedoia
Funding number
AF-ACL consortium; NIH UH3AR076724; and NIH R01AR078762
Project period
Sept 2021- Oct 2022
Principal Investigator
Valentina Pedoia
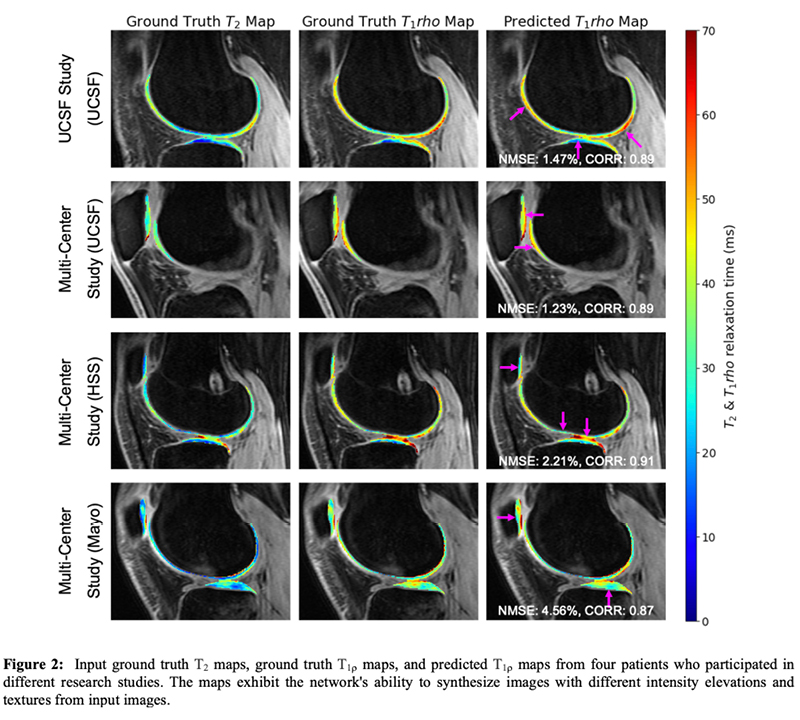
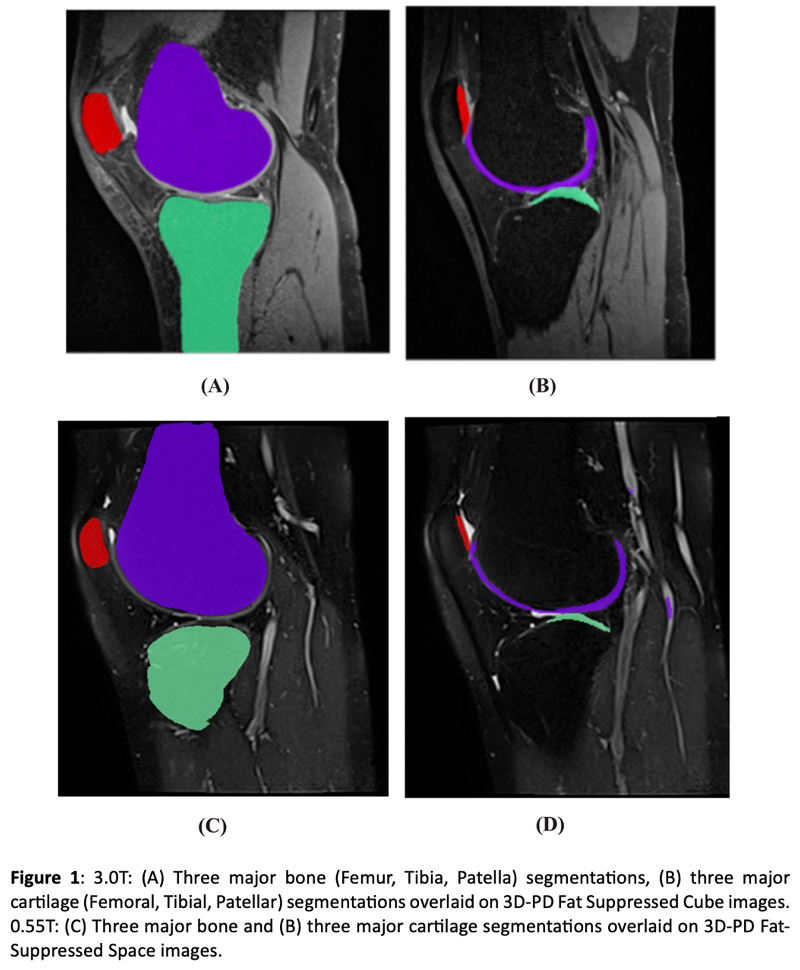
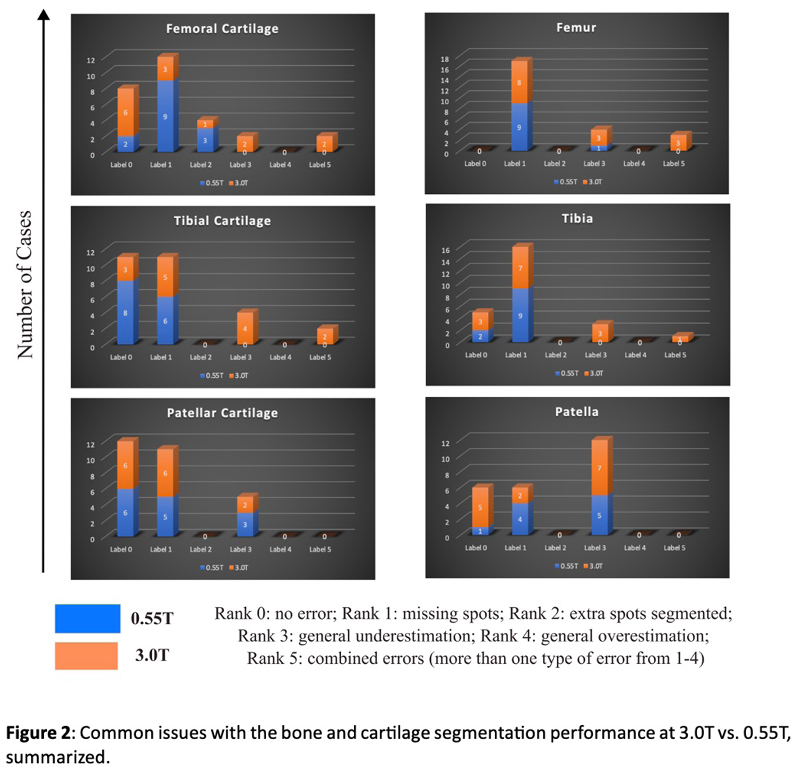
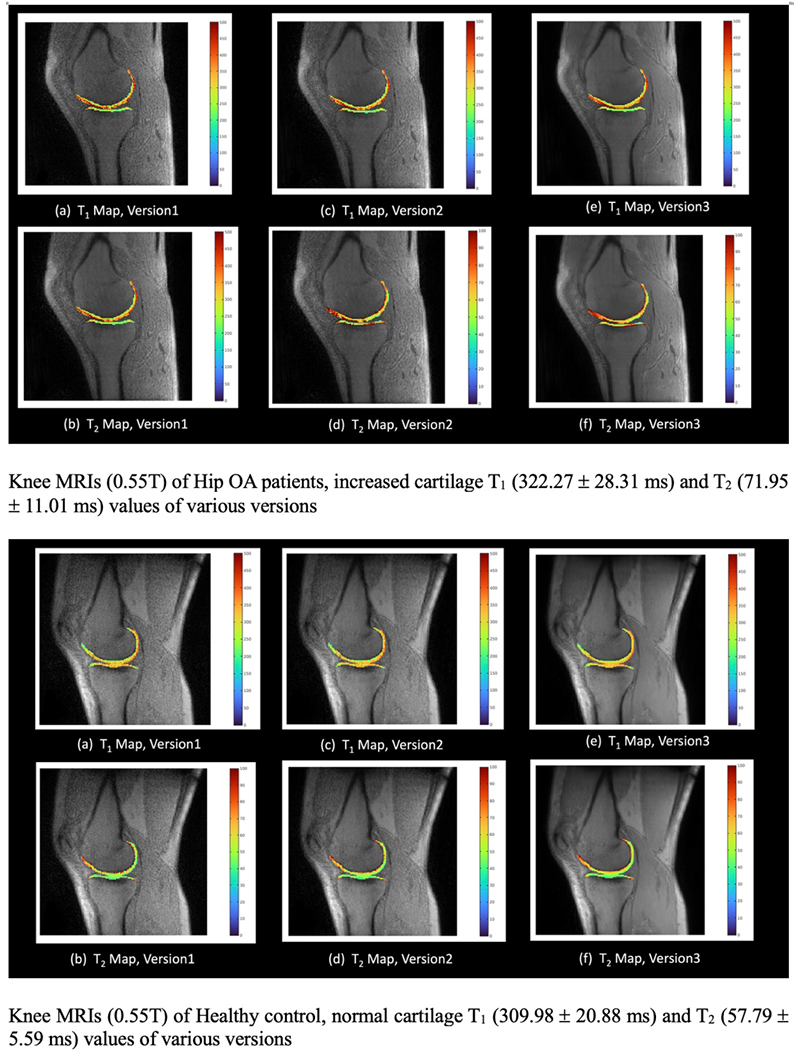
Knee biomarkers in Low-field (0.55T) quantitative MRI: a comparison study w/ 3.0T
Knee biomarkers in Low-field (0.55T) quantitative MRI: a comparison study with 3.0T
In recent years, low-field (0.55T) MRI scanners have experienced a renaissance with novel technical developments ensuring high-quality image acquisition with improved resolution, SNR, and accelerated scan time. Moreover, the 0.55T scanner offers additional advantages such as allowing safer image acquisition due to much lowered specific absorption rates (SAR) and providing higher flexibility to be installed at a broader geography due to lower weight and transportation requirements, with no quench pipe and minimal helium involved. Potential uses of 0.55T scanners have been demonstrated for brain, pulmonary, cardiac, and musculoskeletal imaging domains. Still in its early days, quantitative biomarker estimation and associated reliability have not been explored in-depth for the musculoskeletal system, particularly for knee OA. There is a potential to utilize the existing DL algorithms out-of-the-box, which are extensively validated at higher-field strengths for specific tasks, for application in lower-field. In the current study, our purpose is to evaluate the feasibility of applying deep learning (DL) enabled algorithms to quantify bilateral knee biomarkers in healthy controls scanned at 0.55T and compared with 3.0T; such as Bone Shape, Cartilage Thickness, Compartment-wise cartilage T2 value, etc. We are performing an in-depth comparative analysis of quantitative values between 0.55T vs. 3.0T as well as qualitative evaluations of segmentation quality, performance, and clinical scoring with an MSK-expert radiologist. Overall, using Radial TSE T2 mapping and MR-Fingerprinting T1, T2, and PD mapping methods (in collaboration with Setsompop et al, Stanford University), we are currently evaluating the feasibility of utilizing 0.55T for quantitative parametric evaluation on a varied spectrum of normal to degenerative knee assessment.
Project members at UCSF
Yang Yang, Rupsa Bhattacharjee, Sharmila Majumdar
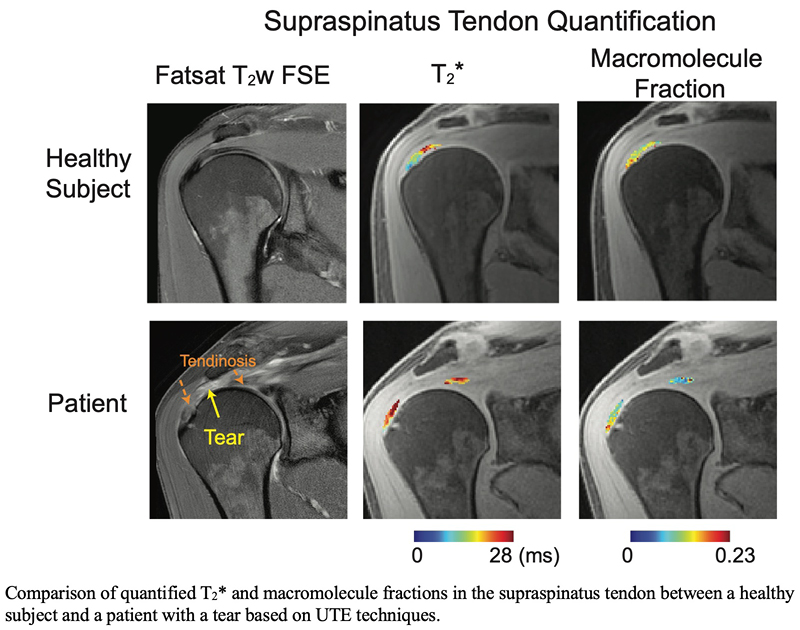
Quantitative MRI to Evaluate Rotator Cuff Tendon Degeneration
Quantitative MRI to Evaluate Rotator Cuff Tendon Degeneration
The primary objectives of this research project are to develop quantitative MRI techniques to assess the quality of tendons and muscles within the rotator cuff. Rotator cuff tears are a common source of pain and disability in the upper extremity. While rotator cuff surgeries are frequently performed, the occurrence of tendon retears poses a significant challenge. We believe the development of accurate quantification methods for both rotator cuff tendons and muscles help to determine optimal treatment strategies. In particular, we are interested in developing ultrashort-echo time MRI techniques for T2* quantification as well as magnetization-transfer quantification.
Related publications
Han M, Larson PE, Link TM, Lansdown DA, Feeley BT, Majumdar S. Quantitative Ultrashort Echo-Time MRI to Assess In Vivo Rotator Cuff Tendon Degeneration. In: Proc the Annual Meeting ISMRM&SMRT, Toronto, ON, Canada, 2023. p. 5113.
Project members at UCSF
Misung Han, Erin Argentieri, Peder Larson, Thomas Link, Drew Lansdown, Brian Feeley, Sharmila Majumdar, Zehra Akkaya, Jocelyn Carpio, Isabelle Remick
Funding number
NIH/NIAMS K01AR075895 (July 2020-June 2025), CTSI RAP grant (July 2021-June 2022)
Principal Investigator
Misung Han (Mentors: Sharmila Majumdar, Brian Feeley, and Peder Larson)
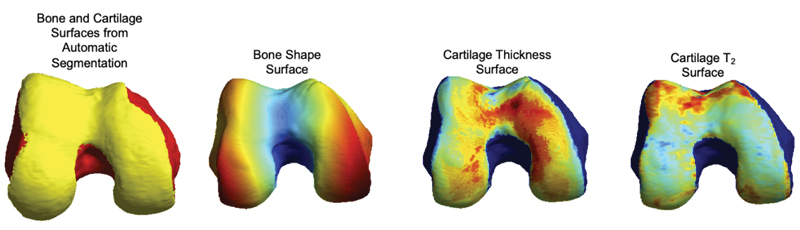
OAI: qMRI 100-D Interpretable Feature Space of Knee OA, A Digital Twin Analysis
Osteoarthritis Initiative (OAI) Project Overview
This extensive body of work is propelled by data-driven methodologies to advance our understanding of osteoarthritis (OA) and its implications for patient outcomes and disease progression. It encompasses various aspects, including disease trajectory analysis, tool development, and prediction techniques, all founded on the extraction of imaging biomarkers through automated deep learning segmentation pipelines. The research aims to inform our understanding of OA through these multifaceted efforts, paving the way for improved patient care within musculoskeletal imaging. This comprehensive approach may encompass diagnosis, prediction, targeted prevention, and personalized treatment strategies, collectively underscoring the transformative potential of data-driven approaches to reshape our comprehension of OA and enhance patient care in the future.
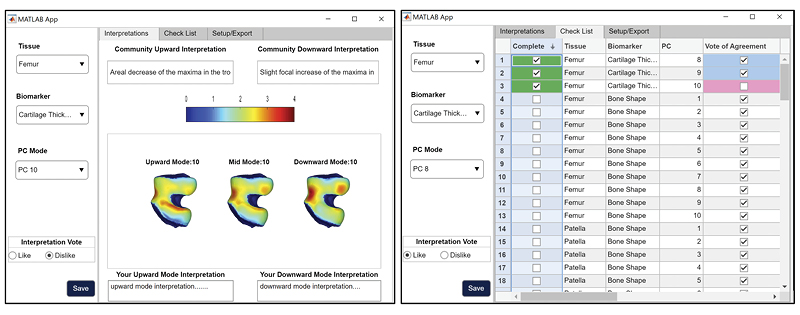
qMRI 100-D Interpretable Feature Space of Knee Osteoarthritis: A Digital Twin Analysis
This multifaceted study, conducted on the Osteoarthritis Initiative (OAI) dataset, represents a significant advancement within the field of musculoskeletal imaging. It encompasses a comprehensive exploration of knee structures, including the femur, patella, tibia, and menisci, while examining a diverse array of imaging biomarkers, such as cartilage thickness, cartilage T2, bone shape, and meniscus shape. Employing cutting-edge techniques like automatic statistical shape modeling and deep learning segmentation pipelines, the study establishes a 100-D interpretable feature space of biomarker-tissue Principal Component (PC) modes. Furthermore, it integrates Digital Twin Analysis, including Image Twin and Clinical Twin analyses, to unveil the intricate relationships between morphological variations and clinical metadata in the context of osteoarthritis (OA) incidence. Through this multidimensional approach, the study provides valuable insights into factors influencing OA development and offers a promising avenue for improving patient care and furthering our understanding of musculoskeletal health.
Related code
https://github.com/gabbieHoyer/OAI-PC-mode-interpreter
Related publications
- Pedoia, Valentina, et al. "Three-Dimensional MRI-Based Statistical Shape Model and Application to a Cohort of Knees with Acute ACL Injury." Osteoarthritis and Cartilage / OARS, Osteoarthritis Research Society, vol. 23, no. 10, 2015, p. 1695, https://doi.org/10.1016/j.joca.2015.05.027.
- Pedoia, V et al. “Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire Osteoarthritis Initiative baseline cohort.” Osteoarthritis and cartilage vol. 27,7 (2019): 1002-1010. doi:10.1016/j.joca.2019.02.800
- Morales Martinez, Alejandro et al. “Learning osteoarthritis imaging biomarkers from bone surface spherical encoding.” Magnetic resonance in medicine vol. 84,4 (2020): 2190-2203. doi:10.1002/mrm.28251
- Iriondo, Claudia et al. “Towards understanding mechanistic subgroups of osteoarthritis: 8-year cartilage thickness trajectory analysis.” Journal of orthopaedic research: official publication of the Orthopaedic Research Society vol. 39,6 (2021): 1305-1317. doi:10.1002/jor.24849
- Morales, Alejandro G et al. “Uncovering associations between data-driven learned qMRI biomarkers and chronic pain.” Scientific reports vol. 11,1 21989. 9 Nov. 2021, doi:10.1038/s41598-021-01111-x
- Xie, E., et al. "Statistical Shape Modeling of the Meniscus from the Osteoarthritis Initiative – A Large-Scale, Data-Driven Evaluation of Demographics and Correlation to Osteoarthritis Incidence." Osteoarthritis and Cartilage, vol. 30, 2022, pp. S276-S277, https://doi.org/10.1016/j.joca.2022.02.373.
- Cummings, Jennifer, et al. "The Knee Connectome: A Novel Tool for Studying Spatiotemporal Change in Cartilage Thickness." Journal of Orthopaedic Research®, https://doi.org/10.1002/jor.25637.
- Gao, Kenneth T., et al. "Large-Scale Analysis of Meniscus Morphology As Risk Factor for Knee Osteoarthritis." Arthritis & Rheumatology, https://doi.org/10.1002/art.42623.
- Hoyer G et al. Quantitative MRI Interpretable 100D Feature Space of Knee Osteoarthritis. In Proceedings of the 31st Annual Meeting of ISMRM, Toronto, Ontario, Canada, 2023. 993.
Project members at UCSF
Gabrielle Hoyer, Jennifer Cummings, Upasana Bharadwaj, Zehra Akkaya, Valentina Pedoia, Sharmila Majumdar
Funding number
Supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), NIH (grant R00-AR-070902).
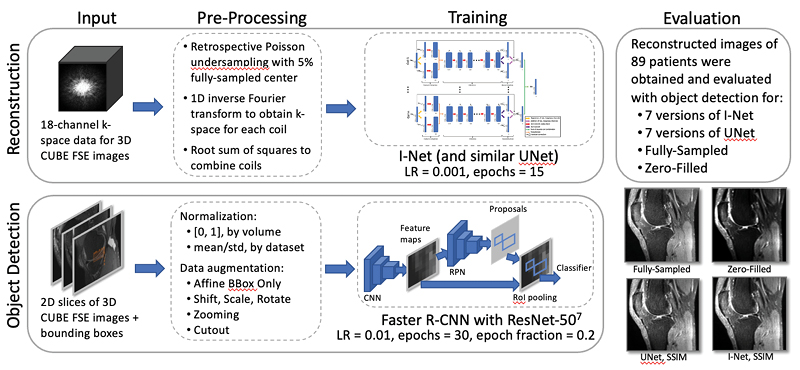
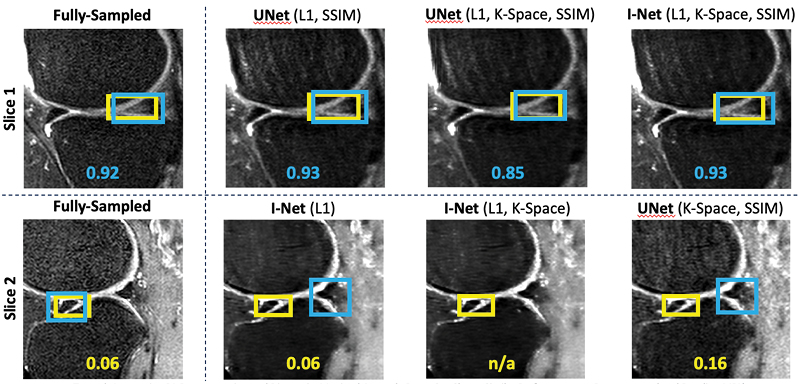
TBrecon: Towards Integration of DL Image Reconstruction & Post-Processing Tasks
TBrecon: Towards Integration of DL Image Reconstruction and Post-Processing Tasks
We are currently working on collecting, annotating, and distributing 3D high-resolution knee MRI data, specifically related to knee osteoarthritis. This dataset includes multi-tissue segmentation and anomaly grading. In our project, we aim to leverage the power of multi-task learning to simultaneously address reconstruction and post-processing tasks within an integrated framework, which encompasses segmentation and object detection. To achieve this, we are developing a strategy based on adversarial robust training and confounding factors learning to ensure the reconstruction of small, rare features, even in under-sampled acquisitions.
Related publications
Konovalova N, Tolpadi A, Liu F, Bhattacharjee R, Gassert F, Giesler P, Luitjens J, Majumdar S, Pedoia V. Towards Integrating DL Reconstruction and Diagnosis: Meniscal Anomaly Detection Shows Similar Performance on Reconstructed and Baseline MRI. In Proceedings of the 31st Annual Meeting of ISMRM, Toronto, Ontario, Canada, 2023. 1381.
Project members at UCSF
Natalia Konovalova, Aniket Tolpadi, Peder Larson, Sharmila Majumdar, Valentina Pedoia
Funding number
NIHR01AR078762 (Apr 1, 2021 - Mar 31, 2026)
Principal Investigator
Valentina Pedoia
People

Staff and Researchers
 Valentina Pedoia, PhD
Valentina Pedoia, PhD
Associate Professor
Joint appointment to Altos Labs as Leader of Image Analysis.
 Misung Han, PhD
Misung Han, PhD
Associate Researcher
Research interest: MRI technical development including acquisition, reconstruction, and quantification. Development of novel and efficient MRI techniques to improve diagnostic capability and to quantitative image biomarkers. Musculoskeletal imaging. Lower back pain. Time-of-flight MR angiography/susceptibility weighted imaging in the brain
 Madeline Hess
Madeline Hess
Research Data Analyst
Research interest: Quantitative imaging, deep learning, computer vision, clinical translation, reproducible and open-source research
Associate Specialist
Research Interest: Osteoarthritis, quantitative MRI, wearable devices/robotics, biomechanics
 Rafeek Thahakoya, PhD
Rafeek Thahakoya, PhD
Postdoctoral Scholar
Research interest: Image acquisition, post processing and analysis of MSK MR images. Development of machine learning tools and quantitative image processing techniques for osteoarthritis conditions of hip and knee joints.
Graduate Student
 Gabrielle Hoyer
Gabrielle Hoyer
PhD Candidate, UC Berkeley-UCSF joint Bioengineering Program
Research interest: Advancing multi-modal machine learning and deep learning, with a dedicated focus on computer vision, computational precision medicine, and digital health. Committed to healthcare innovation, equitable AI, and predictive analysis for global health equity.
 Michelle Tong
Michelle Tong
PhD Student, UC Berkeley-UCSF joint Bioengineering Program
Research interest: Development of robust computer vision, natural language processing, and ML/DL algorithms using medical images and electronic health records. Quantitative biomarker discovery and patient phenotyping for knee osteoarthritis, low back pain, and other musculoskeletal disease. Clinical translational towards improved disease detection and personalized healthcare.
- Erin Argentieri
- Jenn Cummings
Staff
- Steaven Cambell
- Pei Ting Liu
Alumni
Researchers
- Hamza Alizai, MD
- Thomas Baum, MD
- Rupsa Bhattacharjee, PhD
- Radu Bolbos, PhD
- Julio Carballido-Gamio, PhD
- Nathaniel Calixto
- Ko Chiba, MD, PhD
- Alexander Dillon
- Frank Ezekiel
- Jenny Folkesson, PhD
- Matthew Gallo
- Aditi Guha
- Ahi Sema Issever, MD
- Bjorn Jobke, MD
- Pia Jungmann, MD
- Dimitrios Karampinos, PhD
- Subburaj Karupppasamy, PhD
- Martin Kretschmar, MD
- Deepak Kumar, PhD
- Daniel Kuo
- Hans Liebl, MD
- Melissa Lu
- Megan Marsh
- Gerd Melkus, Dr. rer. nat.
- Geetha Mohan, PhD
- Lorenzo Nardo, MD
- Narihiro Okazaki, PhD
- Janina Patsch, MD, PhD
- Jean-Baptiste Pialat, MD
- Prachi Pandit, PhD
- Allison Randolph V
- Colin Russell
- Kiyoshi Sada, PhD
- Benedikt J. Schwaiger, MD
- Dragana Savic, M.Sc and B.Sc.
- Joseph Schooler
- Choongsoo Shin, PhD
- Justin Singh
- Martin Solka
- Carmen Taylor, PhD
- Anand Venkatachari
- Cory Wyatt, PhD
- Zhihong Zhang
- Jin Zuo, PhD
Students
- Fayyaz Ahamed
- Greg Bernstein
- Sarina Bhandari
- Kevin Charette
- Justin Cheung
- Rohit Curucundhi
- Shiktij Dave
- Maxim Daud
- Isabel Dominik
- Jasmine Edelstein
- Charles Fang
- Kevin Gao
- Ayushi Gautam
- Janet Goldenstein
- Riti Gupta
- Tanvi Gupta
- Lindsay Hendrickson
- Madeline Hess
- Jeremy Huang
- Bryce Huerta
- Karim Ibrahim
- Ankita Inamdar
- Gaurav Inamdar
- Nathan Jenkins
- Hannah Jergas
- Solomon Joseph
- Devashish Joshi
- Japdeep Kaur
- Carla Kinnunen
- Joseph Knox
- Christina Kurhanewicz
- Sean Lee
- Erika Legernes
- Taylor Leong
- Lawrence Leung
- Alan Kar-Ming Li
- Trisha Lian
- Katrina Lin
- Lauren Lo
- Kimberly Loo
- Victoria Louie
- George Lu
- Justin Lucas
- Stephen Markham
- Kira McPolin
- Katherine Miclau
- Maya Mundada
- Divya Narayanan
- Hash Nattagh
- Priya Nimbalkar
- Ezinna Nnabue
- Adam Noworolski
- Charles Ouyang
- Alex Pai
- Boyu Pang
- Joyce Pang
- Kayla Panora
- Ishaan Parikh
- Danielle Perez
- Ping Quach
- Sweta Ramachandran
- Adan Romero
- Kiran Sekhon
- Karl Saldanha
- Ehsan Saadat
- Arman Serebrakian
- Swetha Shanbhag
- Sarmad Siddiqui
- Arpita Singhal
- Miki Sode
- Layla Souza
- Derek Speer
- Zoe Swann
- Anna Vardapetyan
- Seth Vigneron
- Yung Wang
- Matthew Zhao
In Memory
Pat Byrd
(1944 - 2010)
Directors
Related Content
- Research Directory
- Abdominal and Pelvic MRI
- Arthritis Imaging Lab (Li Lab)
- Baby Brain Research Group
- Biomagnetic Imaging Laboratory
- Biomechanics & Musculoskeletal Imaging Lab
- Bone Quality Research Lab
- BrainChange Study
- Breast Imaging Research Group
- Breast and Bone Density Group
- Cardiothoracic Imaging Imaging Research
- Center for Molecular and Functional Imaging (CMFI)
- Contrast Material and CT Translational Research Lab
- Evans Lab
- Focused Ultrasound Lab
- High Field MRI Center
- Imaging Research for Neurodevelopment
- Interventional Radiology Research Lab
- Larson Group
- Lupo Lab
- Multimodal Metabolic Brain Imaging
- Musculoskeletal Quantitative Imaging Research
- Neural Connectivity Lab
- Osteoid Osteomas HIFU Clinical Trial
- PET/SPECT Radiochemistry
- PSMA PET Scan
- Physics Research Laboratory
- Program for Molecular Imaging and Targeted Therapy
- Prostate Cancer Imaging Lab (Kurhanewicz)
- Quantitative Biomarkers & AI in MSK Imaging
- Research
- Sarah J. Nelson Lab
- Surbeck Laboratory
- Translational Metabolic Imaging Lab
- Vascular Imaging Research Center
- Viswanath Lab
- Wilson Lab
- Imaging Research Symposium
- Research Conference
- UCSF Radiology at RSNA
- Core Services
MQIR Contact
Mission Bay Campus
1700 4th St., Suite 203
San Francisco, CA 94158
Ph: (415) 514-9655
Fax: (415) 514-9656
China Basin Campus
185 Berry Street, Suite 350
San Francisco, CA 94107
Ph: (415) 353-9401
Fx: (415) 353-9423


