Transfer Learning: Detecting Multiple Sclerosis Lesions with Artificial Intelligence Trained on Other Pathologies
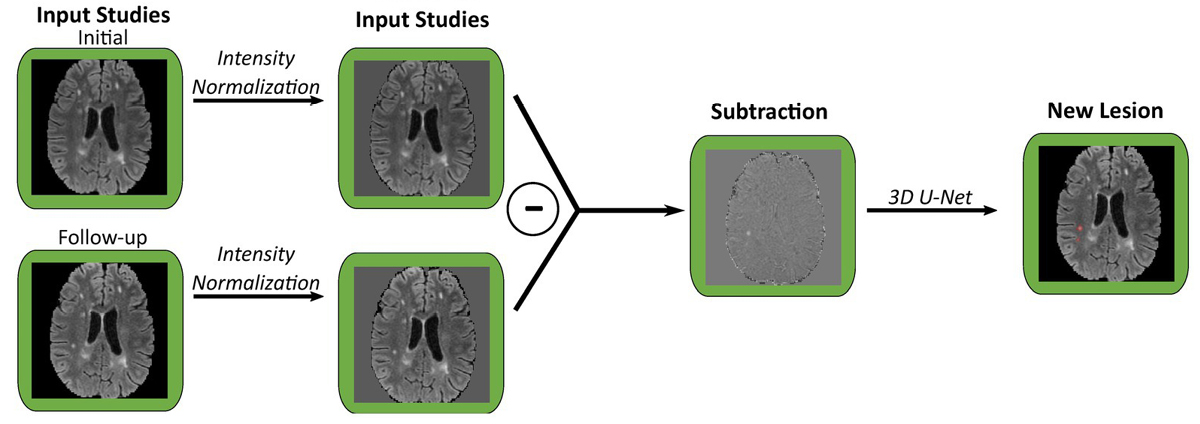
Image credit: Figure 2B, Front. Neurosci., 26 Oct 2023.
When AI leverages large amounts of data, it can be used to produce diagnostic tools that help locate abnormalities faster and more accurately. However, for patients with diseases like multiple sclerosis (MS), the full potential of these new technologies is difficult to achieve, as many published algorithms require a large amount of manually segmented training data that may not be easily available. Even those tools trained on appropriate data sets where they are available can still drop in performance when employed at a new healthcare institution.
UCSF clinicians and imaging scientists have made strides to overcome the hurdle of the large dataset requirement for training deep learning models. In “3D U-Net for automated detection of multiple sclerosis lesions: utility of transfer learning from other pathologies,” published in Frontiers in Neuroscience, the authors explored if models trained on other pathologies can be adapted to identify MS lesions with smaller datasets.
Reflecting on clinical practice, first author Stephen Wahlig, MD, observed that “When interpreting a brain MRI for follow-up of MS, the radiologist’s primary goal is the identification of any new or enhancing (‘active’) lesions. AI algorithms are well-suited to helping radiologists detect MS lesions, but the large data requirement has been prohibitive.”
“While traditionally machine learning algorithms are trained for a particular disease, very different pathologies may have similar imaging features. The algorithm doesn’t know the pathology, and it does not have to – it just needs to learn the features of the lesions it is looking for, and those features might overlap heavily with lesions from other pathologies,” noted Andreas Rauschecker, MD, PhD. “This approach of taking advantage of similarities in imaging features between different pathologies can be used as a general mechanism to decrease the data size burden for training new models and is readily applicable to any pathology.”
Wahlig points to the fact that, “Transfer learning has previously been applied to a variety of other tasks in radiology, ranging from detection of intracranial hemorrhage to classification of fibrotic lung disease.” Applying these insights, Wahlig and the research team modified three existing deep learning models: one for segmenting fluid-attenuated inversion recovery (FLAIR) lesions, one for detecting new FLAIR lesions in follow-up scans compared to baseline, and one for identifying enhancing lesions on T1 post-contrast images.
These models were originally trained on non-MS data such as brain tumors and leukoencephalopathy. The researchers then fine-tuned the models with smaller datasets of MS-specific MRI scans, a technique known as transfer learning. Comparing the performance of fine-tuned models with models trained from scratch (de-novo) on the same MS data and with the original non-MS models, they found that fine-tuning existing models with even a small amount of MS data improved performance. This was particularly evident in positive predictive value (PPV), suggesting that transfer learning reduces the number of false positives.
The study’s results varied depending on the chosen task. Fine-tuning models for single timepoint FLAIR lesion segmentation yielded similar sensitivity to de-novo models but significantly higher PPV, especially with more training data. For uses on new FLAIR lesion segmentation, both fine-tuned and de-novo models outperformed the original model, with fine-tuning showing greater improvement in PPV, though de-novo models had slightly higher sensitivity for some training dataset sizes. However, fine-tuning enhancing lesion segmentation models significantly improved PPV compared to both de-novo models and the original model, with minimal impact on sensitivity.
Transfer learning can be a valuable technique for training deep learning models for MS lesion segmentation, allowing for better performance with smaller datasets compared to training models from scratch. Given that generation of manually segmented data requires a significant investment of time and manpower, this approach could facilitate easier personalization of models for specific institutions.
Rauschecker said, “We hope this strategy of transfer learning from other pathologies, where training data may already be available, can get these AI tools into clinical practice faster – ultimately improving the speed and accuracy with which radiologists review patient’s imaging.”
This study was undertaken by UCSF imaging researchers Stephen Wahlig, MD, Pierre Nedelec, MS , David Weiss, PhD, Jeffrey Rudie, MD, PhD , Leo Sugrue, MD, PhD, and Andreas Rauschecker, MD, PhD.
By Francis Horan
